Sum-Product Networks (SPNs) are a type of probabilistic graphical model (PGMs). They consist of nodes connected by edges and represent distribution functions. In this sense, they do not differ significantly from other PGMs, such as Bayesian or Markov networks. The key difference lies in the semantics of the individual nodes. An SPN is known as a tripartite graph, meaning it has three different types of nodes. Leaf nodes are those in an SPN that have no outgoing edges. These represent univariate distribution functions, i.e., distributions defined over exactly one variable. All internal nodes of an SPN are either sum or product nodes, hence the name Sum-Product Networks. Sum nodes represent mixture distributions, which are a weighted sum of the distributions represented by the directly following nodes, called child nodes. Product nodes represent factorizations, analogous to sum nodes, as the product of the distributions represented by the child nodes.
By combining univariate distributions, factorizations, and mixture distributions, arbitrarily complex distribution functions can be represented. These can be used for various tasks, such as image segmentation, natural language processing, or even in robotics.
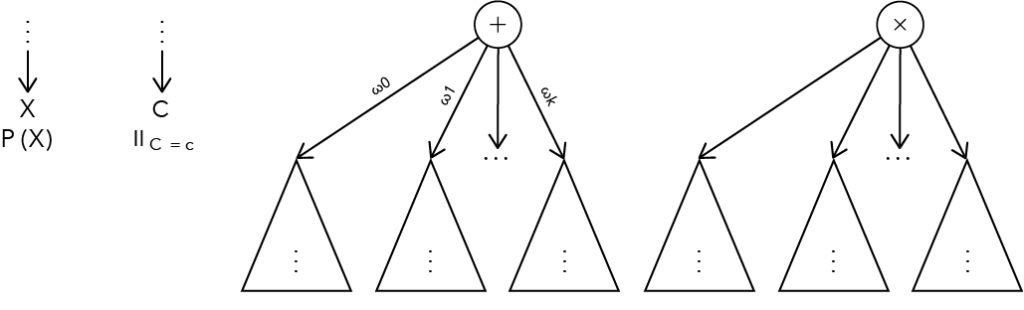
Figure 1: Illustration of the three node types. The two figures on the left represent leaf nodes. A distinction is made between leaf nodes for categorical and continuous variables.
How is a SPN trained?
Training an SPN follows a simple but effective scheme called “LearnSPN,” which learns both the model structure and its parameters. LearnSPN recursively builds the model structure by choosing from five operations to create each node: SplitData, SplitVariables, SplitUninformativeVariables, NaiveFactorization, and CreateLeaf.
SplitData creates a sum node and partitions the dataset using a clustering algorithm like $k$-Means. A sub-SPN is trained recursively for each partition set. These sub-SPNs form the child nodes of the created sum node. The edge weights are chosen proportional to the number of data points in the partition sets, so they sum to 1.
SplitVariables creates a product node and partitions the variables using an independence analysis, such that all variables in one partition set are dependent on each other and as independent as possible from those in the other sets. A sub-SPN is trained for each partition set, considering only the data for the variables in that partition set.
When splitting data and variables, some variables may lose their informative value and become “uninformative,” meaning all data points take the same value for certain variables. In such cases, the SplitUninformativeVariables operation separates these variables by creating a product node with leaf nodes for each uninformative variable. The remaining informative variables are used in a sub-SPN for further training.
A special case of SplitUninformativeVariables is the naive factorization, called NaiveFactorization. Here, a product node is created that receives a leaf node for each variable.
Finally, the CreateLeaf operation creates a leaf node for a variable and estimates a univariate distribution from the given data points. The type of distribution depends on the variable itself.
The decision process that determines which operation to choose is graphically illustrated in Figure 2. It depends on the number of variables $|V|$, the presence of uninformative variables, clustering, and independence, as well as the number of data points $|X|$, which should not fall below a certain threshold $t$. When this threshold is reached, no further splitting of data or variables (only SplitVariables) is performed, ensuring that the number of data points in the leaves is sufficient for a meaningful estimation of the distributions.

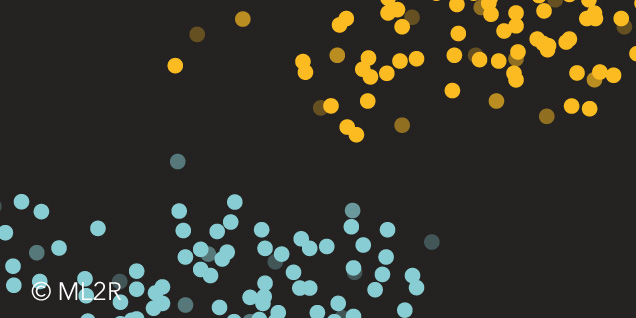
Figure 2: On the left is the decision process during training with LearnSPN and on the right is the training data set consisting of two clusters and an SPN trained on it.
At the root node, the two data clusters (blue and orange) are initially separated. For both clusters, the class membership variable $C$ is then split off, as it has no informative value within a single cluster. Since $C$ is a categorical variable, the corresponding leaves represent indicators for the respective classes. Finally, the two coordinates $X$ and $Y$ are factorized, and normal distributions are estimated from the data.
Inference: Bringing the trained AI model into application
After training an SPN, it is used to predict specific target variables, commonly referred to as inference. In SPN applications, three types of inference are typically distinguished: Maximum a-posteriori (MAP), Most Probable Explanation (MPE), and Maximum Likelihood (MAX).
The fundamental difference between these inference types lies in the division of variables into observed, hidden, and target variables. Observed variables are those that we can observe or use as evidence to predict the target variables. All other variables in our SPN are referred to as hidden variables.
MAP inference seeks the most probable assignment of our target variables given the observed variables (evidence).
If no hidden variables exist, meaning all non-observed variables are target variables, it is called MPE inference, a special case of MAP inference. This type of inference can be used for traditional classification tasks.
MAX inference seeks the most probable assignment of all variables, without distinguishing between variable types.
Let’s consider an example of MPE inference:
First, the values $X=12$ and $Y=12$ are inserted into the appropriate leaf nodes, and the densities are determined according to the estimated distributions. These values are then propagated up to the root node. The path with the highest probability is followed based on the sum node, visiting all target variables. At the corresponding leaf nodes, the assignments for the target variables can be read off. In this case, the highest probability was achieved in the right sub-SPN, and the assignment for $C$ is 1. The point (12, 12) belongs to the orange class 1.
In this way, MPE inference can be computed with just two passes through the graph structure.
Conclusion
In this article, we introduced Sum-Product Networks, a relatively new model class in Machine Learning that, with its clear semantics, efficient inference, and versatile applicability, offers a valuable extension to existing methods. For those interested in exploring SPNs further, we recommend the original work by Poon and Domingos, as well as the review paper by Paris.
